HLA-A29
| HLA-A29 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||||||||
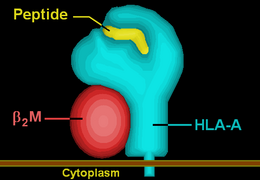
HLA-A29
|
||||||||||||||||
| About | ||||||||||||||||
| Protein | transmembrane receptor/ligand | |||||||||||||||
| Structure | αβ heterodimer | |||||||||||||||
| Subunits | HLA-A*29--, β2-microglobulin | |||||||||||||||
| Older names | A19 | |||||||||||||||
| Subtypes | ||||||||||||||||
|
||||||||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||||||||
|
Subtype
|
allele
|
Available structures
|
|---|---|---|
| A29.1 | *2901 | |
| A29.2 | *2902 | |
| {{{cNick3}}} | *29{{{cAllele3}}} | |
| {{{cNick4}}} | *29{{{cAllele4}}} |
| Study population |
Freq. (in %) |
|---|---|
| Spain Pas Valley | 13.5 |
| Spain Basque Gipuzkoa Pro… | 12.5 |
| South Africa Tswana | 12.2 |
| South African Natal Zulu | 11.0 |
| Cameroon Yaounde | 10.4 |
| Cameroon Pygmy Baka | 10.0 |
| Zimbabwe Harare Shona | 9.6 |
| Burkina Faso Fulani | 9.5 |
| Spain Catalonia Girona | 9.3 |
| Spain Ibizans | 9.3 |
| Tunisia Ghannouch | 9.1 |
| Saudi Arabia | 8.7 |
| Cameroon Bamileke | 8.4 |
| Cameroon Bakola Pygmy | 8.3 |
| Spain Murcia | 8.3 |
| Spain Minorca | 8.2 |
| Spain Basque Arratia Vall… | 8.0 |
| Cameroon Sawa | 7.7 |
| Spain North Cabuernigo | 7.7 |
| Tunisia | 7.2 |
| Portugal Centre | 7.0 |
| Spain Eastern Andalusia | 6.9 |
| Tunisia | 6.7 |
| Cameroon Beti | 6.3 |
| France Corsica | 6.0 |
| Scotland Orkney | 6.0 |
| Spain North Cantabrian | 6.0 |
| Zambia Lusaka | 5.8 |
| Kenya Luo | 5.3 |
| England Liverpool | 5.2 |
| France South East | 5.1 |
| Ireland Northern | 4.9 |
| Kenya | 4.9 |
| Tunisia Tunis | 4.9 |
| USA Hispanic | 4.9 |
| Kenya Nandi | 4.8 |
| Mongolia Hoton | 4.8 |
| Mongolia Khoton Tarialan | 4.8 |
| England Manchester | 4.8 |
| England Newcastle | 4.7 |
| Pakistan Karachi Parsi | 4.4 |
| USA Caucasian (2) | 4.3 |
| Morocco 'berber' Nador Me… | 4.1 |
| Israeli Jews | 3.9 |
| England Lancaster | 3.8 |
| India Jalpaiguri Toto | 3.8 |
| Senegal Niokholo Mandenka | 3.8 |
| Belgium | 3.7 |
| Italy | 3.6 |
| Wales | 3.6 |
| Morocco | 3.4 |
| Mali Bandiagara | 3.3 |
| Ireland South | 3.2 |
| Uganda Kampala | 3.1 |
| Australia New South Wales | 3.0 |
| Brazil | 3.0 |
| Macedonia (4) | 3.0 |
| China Yunnan Han (2) | 2.9 |
| Greece (3) | 2.9 |
| India North Delhi | 2.7 |
| Peru Arequipa | 2.7 |
| Taiwan Hakka | 2.7 |
| USA North American Native… | 2.7 |
| Italy South Campania | 2.6 |
| Italy Bergamo | 2.6 |
| Algeria1 | 2.5 |
| India Kerala Hindu Nair | 2.4 |
| Israel Gaza Palestinians | 2.4 |
| Mexico Mestizos | 2.4 |
| Guatemala Mayans | 2.3 |
| German Essen | 2.0 |
| India West Coast Parsis | 2.0 |
| Israel Arab Druse | 2.0 |
| Romanian | 1.9 |
| Sudanese | 1.8 |
| Pakistan Brahui | 1.7 |
| Thailand Northeast | 1.6 |
| China Harbin Man | 1.5 |
| Guinea Bissau | 1.5 |
| Italy Sardinia pop3 | 1.5 |
| Mexico Sonora Seri | 1.5 |
| Pakistan Pathan | 1.5 |
| Turkey | 1.3 |
| Czech Republic | 1.4 |
| Sweden Stockholm | 1.4 |
| Oman | 1.3 |
| Sweden Uppsala County | 1.2 |
| Russia Nenets | 1.2 |
| Allele frequencies presented, only | |
| Study population |
Freq. (in %) |
|---|---|
| Georgia Tibilisi Kurds | 6.7 |
| Mexico Guadalajara Mestiz… | 6.3 |
| CAR Mbenzele Pygmy | 5.7 |
| Israel Arab Druse | 4.5 |
| Pakistan Karachi Parsi | 4.4 |
| Iran Baloch | 2.8 |
| Taiwan Hakka | 2.7 |
| Kenya | 2.1 |
| Saudi Arabia Guraiat and … | 2.1 |
| India West Coast Parsis | 2.0 |
| India North Hindus | 1.9 |
| Jordan Amman | 1.7 |
| Pakistan Brahui | 1.7 |
| China Guangzhou | 1.5 |
| India New Delhi | 1.5 |
| Pakistan Pathan | 1.5 |
| Singapore Thai | 1.5 |
| South African Natal Zulu | 1.5 |
| Georgia Svaneti Svans | 1.3 |
| Kenya Nandi | 1.3 |
| USA Asian | 1.3 |
| China South Han | 1.2 |
| Finland | 1.1 |
| India North Delhi | 1.1 |
| Kenya Luo | 1.1 |
| China Inner Mongolia | 1.0 |
| Portugal Centre | 1.0 |
| South Korea (3) | 1.0 |
| Pakistan Baloch | 0.8 |
| Croatia | 0.7 |
| Thailand | 0.7 |
| Ireland South | 0.6 |
| Romanian | 0.6 |
| Uganda Kampala | 0.6 |
| USA San Antonio Caucasian… | 0.6 |
| Allele frequencies presented, only | |
HLA-A29 (A29) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α29 subset of HLA-A α-chains. For A29, the alpha "A" chain are encoded by the HLA-A*29 allele group and the β-chain are encoded by B2M . This group currently is dominated by A*2902. A29 and A*29 are almost synonymous in meaning. A29 is a of the serotype A19. A29 is a sister serotype of A30, A31, A32, A33, and A74.
A29 is more common in Western Africa and Southwestern Europe.
For A29-Cw*16-B44 (A*2902:Cw*1601:B*4403) haplotype see Cw*16
...
Wikipedia
