HLA-A3
| HLA-A3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||
HLA-A3
|
||||||||||
| About | ||||||||||
| Protein | transmembrane receptor/ligand | |||||||||
| Structure | αβ heterodimer | |||||||||
| Subunits | HLA-A*03--, β2-microglobulin | |||||||||
| Older names | HL-A3 | |||||||||
| Subtypes | ||||||||||
|
||||||||||
| Rare alleles | ||||||||||
|
||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||
|
Subtype
|
allele
|
Available structures
|
|---|---|---|
| A3.1 | *0301 | |
| A3.2 | *0302 |
|
Subtype
|
allele
|
Available structures
|
|---|---|---|
| A3.5 | *0305 |
| Study population |
Freq. (in %) |
|---|---|
| Finland | 25.0 |
| India Tamil Nadu Nadar | 20.5 |
| Czech Republic | 18.9 |
| Belgium | 17.1 |
| Ireland Northern | 14.3 |
| Australia New South Wales | 13.8 |
| Georgia Svaneti Svans | 13.8 |
| Georgia Tibilisi Georgian… | 13.8 |
| Pakistan Burusho | 13.0 |
| Cape Verde Northwestern I… | 12.0 |
| Ireland South | 11.6 |
| Romanian | 11.6 |
| Croatia | 11.0 |
| USA Caucasians (3) | 10.8 |
| Italy North (1) | 9.7 |
| USA African Americans (2) | 9.4 |
| India Mumbai Marathas | 9.3 |
| India West Bhils | 9.0 |
| Portugal Centre | 9.0 |
| Pakistan Pathan | 8.7 |
| Cameroon Beti | 8.6 |
| Cameroon Bamileke | 7.8 |
| Pakistan Kalash | 7.5 |
| Saudi Arabia Guraiat and … | 7.5 |
| USA Hispanic | 7.3 |
| India North Delhi | 7.2 |
| Madeira | 7.0 |
| Russia Tuva (2) | 6.9 |
| Tunisia | 6.7 |
| USA North American Native… | 6.7 |
| Cape Verde Southeastern I… | 6.5 |
| Azores Santa Maria and Sa… | 6.4 |
| Pakistan Baloch | 6.3 |
| Guinea Bissau | 6.2 |
| Israel Arab Druse | 6.0 |
| South African Natal Zulu | 6.0 |
| USA South Texas Hispanics | 6.0 |
| India North Hindus | 5.8 |
| Zambia Lusaka | 5.8 |
| Iran Baloch | 5.6 |
| Uganda Kampala | 5.5 |
| Jordan Amman | 5.2 |
| Kenya | 5.2 |
| Brazil | 5.1 |
| Oman | 5.1 |
| Cameroon Pygmy Baka | 5.0 |
| Georgia Tibilisi Kurds | 5.0 |
| Sudanese | 4.8 |
| Bulgaria | 4.6 |
| Pakistan Brahui | 4.6 |
| China Beijing | 4.5 |
| Australia Indig. Cape Yor… | 4.4 |
| China Inner Mongolia | 4.4 |
| Mali Bandiagara | 4.4 |
| Pakistan Sindhi | 4.4 |
| China Qinghai Hui | 4.1 |
| Zimbabwe Harare Shona | 4.0 |
| Cameroon Sawa | 3.8 |
| Senegal Niokholo Mandenka | 3.8 |
| Kenya Luo | 3.6 |
| Morocco Nador Metalsa Cla… | 3.4 |
| Kenya Nandi | 3.1 |
| Mex. Guadalajara Mestizos… | 3.1 |
| China Guangzhou | 2.9 |
| China North Han | 2.9 |
| Mongolia Buriat | 2.9 |
| China Tibetans | 2.5 |
| China Yunnan Nu | 2.5 |
| Mexico Mestizos | 2.4 |
| American Samoa | 2.0 |
| Singapore Javanese Indone… | 2.0 |
| Allele frequencies presented, only | |
| Study population |
Freq. (in %) |
|---|---|
| Georgia Tibilisi Kurds | 8.3 |
| Morocco Nador Metalsa Cla… | 4.1 |
| Pakistan Karachi Parsi | 2.8 |
| Israel Arab Druse | 2.5 |
| India New Delhi | 2.3 |
| Sudanese | 2.3 |
| India North Delhi | 2.2 |
| India West Coast Parsis | 2.0 |
| Portugal Centre | 2.0 |
| India Andhra Pradesh Goll… | 1.7 |
| Pakistan Kalash | 1.7 |
| Pakistan Baloch | 1.6 |
| Georgia Tibilisi Georgian… | 1.4 |
| Saudi Arabia Guraiat and … | 1.4 |
| Oman | 1.3 |
| Pakistan Sindhi | 1.3 |
| India North Hindus | 1.0 |
| Pakistan Pathan | 1.0 |
| South Africa Natal Tamil | 1.0 |
| USA South Texas Hispanics | 0.8 |
| Allele frequencies presented, only | |
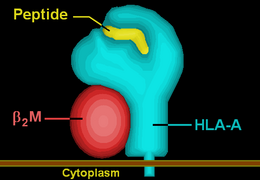
HLA-A3 (A3) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α3 subset of HLA-A α-chains. For A3, the alpha, "A", chain are encoded by the HLA-A*03 allele group and the β-chain are encoded by B2M . This group currently is dominated by A*0301. A3 and A*03 are almost synonymous in meaning. A3 is more common in Europe, it is part of the longest known multigene haplotype, A3-B7-DR15-DQ6.
A3 is primarily composed of A*0301 and *0302 which serotype well with anti-A3 antibodies. There are 26 non-synonymous variants of A*03, 4 nulls, and 22 protein variants.
A3 serotype is a secondary risk factor for myasthenia gravis and lower CD8+ levels in hemochromatosis patients. The HFE (Hemochromatosis) locus lies between A3 and B7 within the A3::DQ6 superhaplotype.
HLA-A3 selects HIV evolution for a mutation Gag KK9 epitope and results in a rapid decline in the CD8 T-cell response. CD8 T-cells are responsible for quickly killing HIV infected CD4+ cells. This type of evolved response may not be specific for HLA-A3 and since HIV is capable of adapting quickly in situ to selective factors.
A*0301 modulates increased risk for Multiple Sclerosis
A3-B7 is part of the A3::DQ2 superhaplotype A3-B8 (Romania, svanS)
A3-B35 (Bulgaria, Croatia, E. Black Sea)
A3-B55 (E. Black Sea)
A3-B7 is bimodal in frequency in Europe with one node in Ireland and the other in Switzerland, relatively speaking Switzerland appears to be higher. A3-Cw7-B7 is one of the most common multigene haplotypes in the western world, particularly in Central and Eastern Europe.
A*0301 : C*0702 : B*0702 : DRB1*1501 : DQA1*0102 : DQB1*0602
...
Wikipedia
